I blogged a couple of months ago about
an imperfect file migration.
One of the reasons this was imperfect (aside from the fact that perhaps all file migrations are imperfect - see below) was because it was an emergency rescue triggered by our Windows 10 upgrade.

Digital preservation is about making best use of your resources to mitigate the most pressing preservation threats and risks. This is a point that Trevor Owens makes very clearly in his excellent new book
The Theory and Craft of Digital Preservation (draft).
I saw an immediate risk and I happened to have available resource (someone working with me on placement), so it seemed a good idea to dive in and take action.
This has led to a slightly back-to-front approach to file migration. We took urgent action and in the following months have had time to reflect, carry out QA and document the significant properties of the files.
Significant properties are the characteristics of a file that should be retained in order to create an adequate representation. We've been thinking about what it is we are really trying to preserve? What are the important features of these documents?
Again, Trevor Owens has some really useful insights on this process and numerous helpful examples in
The Theory and Craft of Digital Preservation. The following is one of my favourite quotes from his book, and is particularly relevant in this context:
“The answer to nearly all-digital preservation question is “it depends.” In almost every case, the details matter. Deciding what matters about an object or a set of objects is largely contingent on what their future use might be.”
So, in fact the title of this blog post is wrong. There is no use in me asking "What are the significant properties of a WordStar file?" - the real question is "What are the significant properties of this particular set of WordStar files from the
Marks and Gran archive?"
To answer this question, a selection of the WordStar files were manually inspected (within a copy of WordStar) to understand how the files were constructed and formatted.
Particular attention was given to how the document was laid out and to the presence of Control and Dot commands. Control commands are markup proceeded by ^ within WordStar - for example ^B to denote bold text). Dot commands are (not surprisingly) proceeded by ‘.’ within WordStar - for example, ‘.OP’ to indicate that page numbering should be omitted within the printed document.
These commands, along with use of carriage returns and horizontal spacing show the intention of the authors.
A few other things have helped with this piece of research.
It is worth also considering the intention of the authors. It seems logical to assume that these documents were created with the intention of printing them out. The use of WordStar was a means to an end - the printed copy to hand out to the actors being the end goal.
I've made the assumption that what we are trying to preserve is the content of the documents in the format that the creator intended,
not the original experience of working with the documents within WordStar.
Properties considered to be significant
The words
Perhaps stating the obvious...but the characters and words present, and their order on the page are the primary intellectual property of this set of documents. This includes the use of upper and lower case. Upper case is typically used to indicate actions or instructions to the actor and also to indicate who is speaking.
It also includes formatting mistakes or typos, for example in some files the character # is used instead of £. # and £ are often confused depending on whether your keyboard is set up as US or UK configuration. This is a problem that people experience today but appears to go back to the mid 1980’s.
Carriage returns
Another key characteristic of the documents is the arrangement of words into paragraphs. This is done in the original document using carriage returns. The screenplays would make little sense without them.
New line for the name of the character.
Blank line.
New line for the dialogue
Another blank line
The carriage returns make the screenplay readable. Without them it is very difficult to follow what is going on, and the look and feel of the documents is entirely different.
Bold text
Some of the text in these files is marked up as bold (using ^B). For example headings at the start of each scene and information on the title page. Bold text is occasionally used for emphasis within the dialogue and thus gives additional information to the reader as to how the text should be delivered: for example “^Bgood^B dog”
Alternate pitch
Alternate pitch is a new concept to me, but it appears in the WordStar files with an opening command of ^A and a closing command of ^N to mark the point at which the formatting should return to ‘Standard pitch’.
The Marks and Gran WordStar files appear make use of this command to emphasise particular sections of text. For example, one character tells another that he has seen Hubert with “^Aanother woman^N”. The fact that these words are displayed differently on the page, gives the actor additional instruction as to how these words should be spoken.
The
Wordstar 4.0 manual describes how alternate pitch relates to the character width that is used when the file is printed and that “WordStar has a default alternate pitch of 12 characters per inch”.
However, in a printed physical script that was located for reference (which appears to correspond to some of the files in the digital archive), it appears that text marked as alternate pitch was printed in italics. We can not be sure that this would always be the case.
However this may be interpreted, the most important point is that Marks and Gran wanted these sections of text to stand out in some way and this is therefore a property that is significant.
Underlined text
In a small number of documents there is underlined text (marked up with ^S in WordStar). This is typically used for titles and headings.
As well as being marked up with ^S, underlined text in the documents typically has underscores instead of spaces. This is no doubt because (as described in the manual), spaces between underlined words are not underlined when you print. Using underscores presumably ensures that spaces are also underlined without impacting on the meaning of the text.
Monospace font
Although font type can make a substantial difference to the layout of a page, the concept of font (as we understand it today) does not seem to be a property of the WordStar 4.0 files themselves, however I do think we can comfortably say that the files were designed to be printed using a monospace font.
WordStar itself does not provide the ability to assign a specific font, and the fact that the interface is not WYSIWYG means font can not be assumed by seeing the document in a native environment.
Searching for 'font' within the WordStar manual brings up references to 'italics font' for example but not modern font type as we know it. It does however talk about using the .PS command to change to 'proportional spacing'. As described in the
manual:
"Proportional spacing means that each character is allocated space that is proportional to the character's actual width. For example, an i is narrower than an m, so it is allocated less horizontal space in the line when printed. In monospacing (nonproportional spacing), all characters are allocated the same horizontal space regardless of the actual character width."
The .PS command is not used in the Marks and Gran WordStar files so we can assume that monospace font is used.
This is backed up by looking at the physical screenplays that we have in the Marks and Gran archive. The font on contemporary physical items is a serif font that looks similar to Courier.
This is also consistent with the description of screenplays as described on wikipedia: “The standard font is 12 point, 10 pitch Courier Typeface”.
Courier font is also mentioned in the description of a WordStar migration by
Jay Gattuso and Peter McKinney (2014).
Hard page breaks
The Marks and Gran WordStar files make frequent use of hard page breaks. In a small selection of files that were inspected in detail, 65% of pages ended with a hard page break. A hard page break is visible in the WordStar file as the .PA command that appears at the bottom of the page.
As described in the
wikipedia page on screenplays “The format is structured so that one page equates to roughly one minute of screen time, though this is only used as a ballpark estimate”.
This may help explain the frequent use of hard page breaks in these documents. As this is a deliberate action and impacts on the look of the final screenplay this is a property that is considered significant.
Text justification
In most of the documents, the authors have positioned the text in a particular way on the page, centering the headings and indenting the text so it sits towards the right of the page. In many documents, the name of a character sits on a new line and is centred, and the actual dialogue appears below. This formatting is deliberate and impacts on the look and feel of the document thus is considered a significant property.
Page numbering
Page numbering is another feature that was deliberately controlled by the document creators.
Many documents start with the .OP command that means ‘omit page numbering’.
In some documents page numbering is deliberately started at a later point in the document (after the title pages) with the .PN1 command to indicate (in this instance) that the page numbering should start at this point with page 1.
Screenplay files in this archive are characteristically split into several files (as is the recommended practice for longer documents created in WordStar). As these separate files are intended to be combined into a single document once printed, the inclusion of page numbers would have been helpful. In some cases Marks and Gran have deliberately edited the starting page number for each individual file to ensure that the order of the final screenplay is clear. For example the file CRIME5 starts with .PN31 (the first 30 pages clearly being in files CRIME1 to CRIME4).
Number of pages
The number of pages is considered to be significant for this collection of WordStar files. This is because of the way that Marks and Gran made frequent use of hard page breaks to control how much text appeared on each page and occasionally used the page numbering command in WordStar.
Note however that this is not an exact science given that other properties (for example font type and font size) also have an impact on how much text is included on each page.
Just to go back to my previous point that the question in the title of this blog is not really valid...
Other work that has been carried out on the preservation of a collection of WordStar files at the National Library of New Zealand reached a different conclusion about the number of pages. As described by
Jay Gattuso and Peter McKinney, the documents they were working with were not screenplays, they were oral history transcripts and they describe one of their decisions below:
"We had to consider therefore if people had referenced these documents and how they did so. Did they (or would they in future) reference by page number? The decision was made that in this case, the movement of text across pages was allowable as accurate reference would be made through timepoints noted in the text rather than page numbers. However, it was an impact that required some considerable attention."
Different type of content = different decisions.
Headers
Several documents make use of a document header that defines the text that should appear at the top of every document in the printed copy. Sometimes the information in the header is not included elsewhere in the document and provides valuable metadata - for example the fact that a file is described in the header as "REVISED SECOND DRAFT” is potentially very useful to future users of the resource so this information (and ideally it's placement within the header of the documents as appropriate) should be retained.
Corruption
This is an interesting one. Can corruption be considered to be a significant property of a file? I think perhaps it can.
One of the 19 disks from the Marks and Gran digital archive appears to have suffered from some sort of corruption at some stage in its life. Five of the files on this disk display a jumble of apparently meaningless characters at one or more points within the text. This behaviour has not been noted on any of the other files on the other disks.
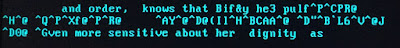
The corruption can not be fixed. The original content that has been lost can not be replaced. It therefore needs to be retained in some form.
There is a question around how this corruption is presented to future users of the digital archive. It should be clear that some content is missing because corruption has occurred, but it is not essential that the exact manifestation of the corruption is preserved in access copies. Perhaps a note along the lines of ...
[THE FILE WAS CORRUPT AT THIS POINT. SOME CONTENT IS NO LONGER AVAILABLE]
...would be helpful?
Very interested to hear how others have dealt with this issue.
Properties not considered to be significant:
Other properties noted within the document were thought to be less significant and are described below:
Font size
The size of a font will have a substantial impact on the layout and pagination of a document. This appears to have been controlled using the Character Width (.CW) command as described in the manual:
"In WordStar, the default character width is 12/120 inch. To change character width, use the dot command .CW followed by the new width in 120ths of an inch. For example, the 12/120 inch default is written as .CW 12. This is 10 characters per inch, which is normal pitch for pica type. "
The documents I'm working with do not use the .CW command so will accept the defaults. Trying to work out what this actually means in modern font sizes is making my head hurt. Help needed!
As mentioned above, the description of
screenplays on wikipedia states that: “The standard font is 12 point, 10 pitch Courier Typeface”. We could use this as a guide but can't always be sure that this standard was followed.
In the
National Library of New Zealand WordStar migration the original font is considered to be 10 point Courier.
Word wrap
Where hard carriage returns are not used to denote a new line, WordStar will wrap the text onto a new line as appropriate.
As this operation was outside the control of the document creator, this property isn’t considered to be significant. This decision is also documented by the National Library of New Zealand in their work with WordStar files as discussed in
Gattuso and McKinney (2014).
Soft page breaks
Where hard page breaks are not used, text flows on to the next page automatically.
As this operation was not directly controlled by the document creator it is not considered to be significant.
In conclusion
Defining significant properties is not an exact science, particularly given that properties are often interlinked. Note, that I have considered number of pages to be significant but other factors such as font size, word wrap and soft page breaks (that will clearly influence the number of pages) to be not so significant. Perhaps there is a flaw in my approach but I'm running with this for the time being!
This is a work in progress and comments and thoughts are very welcome.
I hope to blog another time about how these properties are (or are not) being preserved in the migrated files.
Jenny Mitcham, Digital Archivist